The Development of a Multiplex Reverse Transcription Polymerase Chain Reaction for Detection of Porcine Reproductive and Respiratory 
 Author
Author  Correspondence author
Correspondence author
Genomics and Applied Biology, 2010, Vol. 1, No. 4 doi: 10.5376/gab.2010.01.0004
Received: 18 Nov., 2010 Accepted: 10 Dec., 2010 Published: 24 Dec., 2010
Wu et al., 2010, The Development of a Multiplex Reverse Transcription Polymerase Chain Reaction for Detection of Porcine Reproductive and Respiratory Syndrome Virus, Genomics and Applied Biology, 2010, Vol. 1, No. 4 (DOI: 10.5376/gab.2010.01.0004)
A multiplex reverse transcription polymerase chain reaction (multiplex RT-PCR) was developed for the detection of porcine reproductive and respiratory syndrome virus (PRRSV). A set of three pairs of primer were designed based on the sequence of high conservative region of PRRSV. The diagnostic accuracy of the multiplex RT-PCR assay was evaluated using 25 field clinical samples, 10 reference strains and 6 true-negative samples. Simultaneously, the specificity and sensitivity of this method were evaluated. The result indicated that this assay could reliably differentiate between PRRSV and other swine viral disease, such as classical swine fever virus (CSFV), swine vesicular disease virus (SVDV) and vesicular exanthema of swine virus (VESV). Meanwhile, this assay was shown to be 10-fold more sensitive than the conventional single RT-PCR (conventional sRT-PCR) method. The results indicated that the multiplex RT-PCR developed in this study has high sensitivity, strong specificity and easy operation,it is not only suitable for diagnosis of samples of insignifiance content,but also it has important applied value in getting rid of animals with recessive virus and veterinary quarantine .The method may provide a new avenue to the rapid detection of this important pathogen in one reaction.
Porcine reproductive and respiratory syndrome virus (PRRSV) is a member of the family Arteriviridae (Cavanagh, 1997). It is characterized by respiratory disease in young pigs and severe reproductive failure in sows, including abortion, stillbirths and weak piglets (Hill, 1990). PRRSV has caused immense economic losses in the pig industry and is considered to be one of the most important infectious diseases of pigs in the world (Polson et al., 1992; Li et al., 2007), and it is comprised of two viral genotypes, the North American type and the European type (NA-type and EU-type) (Nelsen et al., 1999). An et al (2007) found that all of the Chinese isolates examined belong to the North American (NA) type and the viruses were geographically restricted to regions in southeast China. The virus was first confirmed in China in 1996, since then, the virus has spread widely in China (Gao et al., 2004; Chen et al., 2006). In June of 2006, outbreaks of highly pathogenic (acute, atypical) PRRS in most areas of China have been suggested to be caused by highly virulent Chinese-type PRRSV (H-PRRSV) strains. From January to July 2007, 39 455 morbid pigs died among 143 221 infected pigs according to the government numerical data. Rapid spread and their persistence in some environments have made the control of outbreaks difficult and, at times, even impossible. Therefore, development of a rapid, sensitive and specific diagnostic method is extremely important to avoid or prevent the occurrence and spread of this disease.
Due to the highly variability of PRRSV, three sets of primers in different region of PRRSV genome were designed would improve the detection sensitivity of PRRSV. In the test, if one or more fragment was amplified, but negative control didn't amplify any fragment, which indicated that PRRSV existed in the samples. A conventional single RT-PCR (sRT-PCR) assay for PRRSV is commercially available in China and this technique has provided a simple and rapid technique to detect clinically suspicious samples. However, this method is unable to detect virus in serum and blood in which the amount of PRRSV is very low. To obtain a better detection method for PRRSV, in this study three sets of primers were designed and used in a multiplex RT-PCR format to detect in a number of tissues and organs, including serum, blood, heart, brain, lung and kidney. Meanwhile, as a RNA virus, one-step multi-RT-PCR was developed will detect suspected samples, whitch has certain instructive significance to preventive treatment of PRRSV in production practice extensively.
1 Results and Analysis
1.1 The sensitivity of CH-1a RNA detection by the multiplex RT-PCR and the conventional RT-PCR
The sensitivity of multiplex RT-PCR reaction was compared with RT-PCR reaction for detecting PRRSV using 10-fold serial dilutions Marc-145 cell-adapted cultures infected by CH-1a live virus. All ampliï¬ed DNA products were electrophoresed on 1.5% agarose gel stained with ethidium bromide. sRT-PCR reaction had a detection limit of 0.1 TCID50 while multiplex RT-PCR reaction had a detection limit of 0.01 TCID50 (Figure 1). Therefore, the sensitivity of multiplex RT-PCR reaction for detecting PRRSV is about 10 times higher than that of sRT-PCR reaction. As shown in Figure 1, the multiplex RT-PCR could detect the presence of the virus up to the 10−7 dilution whereas the conventional sRT-PCR was only able to detect up to the 10−6 dilution. Therefore, the detection limit or sensitivity of the multiplex RT-PCR was at least 10 fold higher than that of the conventional sRT-PCR.
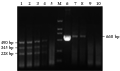 Figure 1 Sensitivity of CH-1a RNA detection by the multiplex RT-PCR and the conventional RT-PCR |
1.2 The speciï¬city test for the multiplex RT-PCR
The multiplex RT-PCR was facilitated to amplify the reference virus strain of one SVDV sample (HK/70), two CSFV samples (C-strain and Shimen virus) and two VESV isolates (serotypes B51 and H54). None of the CSFV, SVDV and VESV isolates was positive by the multiplex RT-PCR (Figure 2).This results showed that the multiplex RT-PCR is specific for PRRSV.
 Figure 2 The result of the speciï¬city test for the multiplex RT-PCR. Three related viruses (CSFV, SVDV, VESV) were assayed, and all of them were negative |
1.3 The accuracy evaluation of the multiplex RT-PCR assay
The accuracy of the multiplex RT-PCR assay was evaluated by detection of 25 field clinical samples (collected from Guangxi, Gansu and Ningxia province of China), 10 reference strains and 6 true-negative samples. The coincidence of the multiplex RT-PCR and the conventional sRT-PCR was 100% for 10 reference strains and 6 true-negative samples (As Table 1 showed). With regards to the 25 field clinical samples, 19 samples were detected positive by the multiplex RT-PCR assay, 15 samples by the conventional RT-PCR assay. Notably, all sample detected positive by the conventional RT-PCR were positive by the multiplex RT-PCR. Meanwhile, sequence analysis and virus isolation were verified this results (Table 2). This indicated that the sensitivity of the multiplex RT-PCR for PRRSV was higher than the conventional sRT-PCR.
| Table 1 The coincidence of the multiplex RT-PCR and the conventional RT-PCR by detection of the PRRSV reference samples |
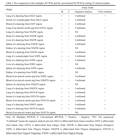 Table 2 The comparison of the multiplex RT-PCR and the conventional RT-PCR by testing 25 clinical samples |
1.4 The feasibility analysis of the multiplex RT-PCR during the course of PRRSV infection
In order to assess the performance of the multiplex RT-PCR during the course of PRRSV infection, three pigs were experimentally infected with PRRSV HuN with 1.5 mil the titers of 105.8 TCID50 by dropped nose (denoted No.1, No.2 and No.3). Blood samples were collected over a 20 day period and tested using the multiplex RT-PCR and the conventional RT-PCR. One uninfected control pig was also included as a negative control. All of the samples were positive from the sixth to the fourteenth day after challenge by both PCR methods. All of the samples were positive from sixth to fourteenth day after challenge by both PCR methods. However, one sample on the fourth day and two samples on the sixteenth day were only detected positive by the multiplex RT-PCR, indicating the ability of the multiplex RT-PCR for detection of low concentration virus. One negative control from, tissue of a healthy swine was tested negatively during the test process (Table 3).
 Table 3 Comprison between the multiplex RT-PCR (M) and the conventional RT-PCR (C) for detection of PRRSV blood samples from swine artificial infected |
2 Discussion
Multiplex PCR is gaining popularity because of its experimental simplicity and greater effectiveness as well as decreased effort and shorter time required (Chamblain et al., 1998). Meanwhile, this method were be used to detect FMDV were confirmed 100 times more than sRT-PCR (Bao et al., 2008).However, the care still needs to be taken to overcome the often problems such as a number of primers used in a same reaction tube might interact with each other, leading to the block of the reaction (Elnifro et al., 2000; Henegariu et al., 1997; Markoulatos et al., 2002) and close size of PCR products or cross reactions, leading to complex for distinguish the origin of the PCR products. Therefore, much care has been taken to select and design the multiplex RT-PCR primers in this study so that the obtained PCR products could be very specific and easily identified by separating the products onto agarose gel by electrophoresis.
We showed that the multiplex RT-PCR method was approximately 10 times more sensitive than the standard conventional sRT-PCR. This makes the multiplex RT-PCR a better choice over the conventional sRT-PCR for the diagnosis of PRRSV in case examination, especially when the lower concentration of virus. Therefore, the method has a potential advantage for detection of the virus from serum, blood, spleen, asymptomatic carriers and co-infection cases with other virus. Because of using of one-step RT-PCR method in this multiplex RT-PCR, more time was saved in this assay. A standard two-step PCR takes at least 3~4 h whereas the one-step multiplex RT-PCR reaction requires 2~3 h including time for identification of the PCR products by gel electrophoresis. Moreover, the one-step multiplex RT-PCR takes RNA as templates, which not only saved the time of cDNA synthesis required in the conventional sRT-PCR, but also decreased the possibility that losing the yield of RNA during the cDNA synthesis process. This is because that in multiplex RT-PCR, the cDNA synthesis and ampliï¬cation are achieved at the same time in a single tube. The merits of this method suggested that it is suitable molecular method for the rapid, accurate and reliable detection of PRRSV in clinical samples. In particular, when distinguishing of swine viruses (CSFV, SVDV and VESV) is required.
Considering the prevalence and economic impact of PRRSV, a simple, cost effective, sensitive and rapid diagnostic technique is urgently required. Although the multiplex RT-PCR method described in this study showed some advantages over the conventional sRT-PCR, further evaluation of this assay on a larger scale is strongly recommended.
3 Materials and Methods
3.1 Reference strains and samples
CH-1a (GenBank accession number: AY032626) live vaccine virus and other reference strains (Table 4) used in this study was obtained from the virology department of Lanzhou Veterinary Research Institute, Gansu of P.R. China. Six true-negative samples were collected from healthy animals. Twenty-five clinical samples from 25 pigs, including lungs, livers, serums and other organs, were obtained from the pigs that were suspected with the infection of PRRSV and stored at −70℃ until total RNA extraction. Ten blood samples from three experimental PRRSV-infected swine and one healthy control pig were also included in the study.
.png) Table 4 10 reference strains used in this study |
3.2 Virus isolation
The samples were processed as described elsewhere (Scortti M et al., 2006), and used to inoculate monolayers of Marc-145 cells in duplicate. These cells were incubated for 90 min at 37℃ to facilitate adsorption. They were washed twice with DMEM( Dulbecco's Modified Eagle Medium) and we then added fresh DMEM supplemented with 10% fetal bovine serum (FBS) (HyClone, USA) and incubated the cells for 6 days at 37℃, in a humidiï¬ed atmosphere containing 5% CO2. With Cells were examined for cytopathic effect (CPE) on days 4~6. HuN strain cell virus were be used as positive control. Virus-free DMEM or FBS were used as negative controls. If CPE was observed, RT-PCR was carried out to confirm the presence of PRRSV.
3.3 Extraction of RNA
The viral RNA of all clinical samples were extracted using QIA amp Viral RNeasy Mini Kit (Qiagen, Germany), following manufacturer’s instructions. In brief, after lysis of the specimens, the mixture was applied to a spin column as described by the manufacturer’s protocol. The extracted RNA was eluted in a total volume of 60 μL of elution buffer and was stored at −70℃ until used.
3.4 Primers design
Based on the conserved nucleotide sequence of the PRRSV, three pairs of PRRSV specific primers were designed using DNAStar software( DNAstar 7.0 green.rar )and the sequence of the primers, their position on the PRRSV genome and the expected product size are shown in Table 5.
 Table 5 Specific primer pairs used for the multiplex RT-PCR |
3.5 RT-PCR reaction
Conventional sRT-PCR was performed using a commercially available two-step assay purchased from Guangdong Biological Technology Co. Ltd (China) following the manufacturer's instructions. The one-step multiplex RT-PCR was performed using TaKaRa one-step RT-PCR kit (TaKaRa Biotechnology Co., Ltd, China). Three pairs of primers were employed in one reaction. The multiplex RT-PCR and the conventional single RT-PCR (sRT-PCR) were compared at the same time by testing each sample. The multiplex RT-PCR was carried out in a reaction volume of 50 μL containing 0.5 μL of each primer (12.5 pmol/μL), 5 μL of 10× one step RT-PCR Buffer, 5 μL 2.5 mmol/L dNTPs, 10 μL of 25 mml/L MgCl2, 1 μL of RNase Inhibitor (40 U/μL), 1 μL of AMV Reverse Transcriptase XL (5 U/μL), 1 μL of AMV-optimized Taq (5 U/μL) and 10 μL of each viral RNA. Amplification conditions were an incubation at 50℃ for 30 min and an initial denaturing at 94℃ for 2 min, and then followed by 30 cycles, which consisted of denaturing at 94℃ for 40 s, annealing at 57℃ for 40 s, and extension at 72℃ for 50 s. The last extension step was performed at 72℃ for 8 mins, and then kept the reaction at 4℃. Reaction products were analyzed by agarose gel electrophoresis and stained with ethidium bromide.
3.6 Sensitivity and specificity test
The sensitivity of the multiplex RT-PCR assay was determined using 10-fold serial Marc-145 cell-adapted cultures infected by CH-1a live virus with the titer of 105.0 TCID50. The specificity of the multiplex RT-PCR was determined by testing different non-PRRSV isolates related to high fever (≥40.5℃) and reddened skin (including classical swine fever virus (CSFV), swine vesicular disease virus (SVDV ) and vesicular exanthema of swine virus (VESV).
3.7 Sequencing
The purified multiplex RT-PCR products (490 bp) and the conventional sRT-PCR products (660 bp) by the gel of 25 clinical samples were subjected to sequencing (Takara, Dalian, China). The sequences were analyzed using the DNAStar to confirm the correction of the amplifications.
Authors' contributions
Conceived the experiment and edited the manuscript: Liu Xiangtao and Wu Jinyan. Data collection and analysis: Tian Hong Chen Yan Shang Youjun Yin Shuanghui Zhao Na Jin Ye and Xie Qingge. Wrote the manuscript: Wu Jinyan. All authors have read the final version of this paper and agreed with the authors’ credits.
Acknowledgements
This work was supported by the national “863” Key Technology R&D Programme (Grant No. 2006AA241110).
References
An T., Zhou Y., Liu G., Tian Z., Li J., Qiu H., and Tong G., 2007, Genetic diversity and phylogenetic analysis of glycoprotein 5 of PRRSV isolates in mainland China from 1996 to 2006: Coexistence of two NA-subgenotypes with great diversity, Vet. Mic., 123(1-3): 43-52 doi:10.1016/j.vetmic.2007.02.025 PMid:17383833
Bao H.F., Li D., Guo J.H., Lu Z.J., Chen Y.L., Liu Z.X., Liu X.T., and Xie Q.G., 2008, A highly sensitive and speciï¬c multiplex RT-PCR to detect foot-and-mouth disease virus in tissue and food samples, Arch. Virol., 153(1): 205-209 doi:10.1007/s00705-007-1082-y PMid:17987350
Cavanagh D., 1997, Nidovirales: a new order comprising Coronaviridae and Arteriviridae, Arch. Virol., 142(3): 629-633 PMid:9349308
Chamblain J.S., Gibbs R.A., Renier I.E., Nguyen P.N., and Caskey C.T., 1998,Deletion screening of the duchenne muscular dystrophy locus via multiplex DNA ampliï¬cation, Nucl. Acids. Res., 16(23): 11141-11156 doi:10.1093/nar/16.23.11141 PMid:3205741 PMCid:339001
Chen J., Liu T., Zhu C.G., Jin Y.F., and Zhang Y.Z., 2006, Genetic variation of Chinese PRRSV strains based on ORF5 sequence, Biochem. Genet., 44(9-10): 421-431 doi:10.1007/s10528-006-9039-9 PMid:17048090
Elnifro E.M., Ashshi A.M., Cooper R.J., and Klapper P.E., 2000, Multiplex PCR: Optimization and application in diagnostic virology, Clin. Microbiol. Rev.,13(4): 559-570 doi:10.1128/CMR.13.4.559-570.2000 PMid:11023957 PMCid:88949
Gao Z.Q., Guo X., and Yang H.C, 2004, Genomic characterization of two Chinese isolates of porcine respiratory and reproductive syndrome virus, Arch. Virol., 149(7): 1341-1351 doi:10.1007/s00705-004-0292-0 PMid:15221535
Henegariu O., Heerema N.A., Dlouhy S.R., Vance G.H., and Vogt P.H., 1997, Multiplex PCR: Critical parameters and step-by-step protocol, Biotechniques, 23: 504-511 PMid:9298224
Hill H., 1990, Overview and history of mystery swine disease (swine infertility and respiratory syndrome), In: Proceedings of the Mystery Swine Disease Communication Meeting, Denver, CO., pp.29-31
Li Y.F., Wang X.L., Bo K.T., Wang X.W., Tang B., Yang B.S., Jiang W.M., and Jiang P., 2007, Emergence of a highly pathogenic porcine reproductive and respiratory syndrome virus in the Mid-Eastern region of China, Vet. J., 174(3): 577-584 doi:10.1016/j.tvjl.2007.07.032 PMid:17869553
Markoulatos P., Mangana-Vougiouka O., Koptopoulos G., Nomikou K., and Papadopoulos O., 2002, Detection of sheep poxvirus in skin biopsy samples by a multiplex polymerase chain reaction, J. Virol. Methods, 84(2): 161-167 doi:10.1016/S0166-0934(99)00141-X
Nelsen C.J., Murtaugh M.P., and Faaberg K.S., 1999, Porcine reproductive and respiratory syndrome virus comparison: divergent evolution on two continents, J. Virol., 73(1): 270-280
PMid:9847330 PMCid:103831
Polson D.D., Marsh W.E., and Dial G.D., 1992, Financial evaluation and decision making in the swine breeding herd, Veterinary Clinics of North America Food Animal Practice 8(3): 725-747
PMid:1446279
Scortti M., Prieto C., Martınez-Lobo F.J., Simarro I., and Castro J.M., 2006, Effects of two commercial European modiï¬ed-live vaccines against porcine reproductive and respiratory syndrome virusesin pregnant gilts, Vet J., 172(3): 504-514 doi:10.1016/j.tvjl.2005.07.015 PMid:16169756
Suarez P., Zardoya R., Prieto C., Solana A., Tabares E., and Bautista J.M., 1994, Direct detection of the porcine reproductive and respiratory syndrome (PRRS) virus by reverse polymerase chain reaction (RT-PCR), Arch. Virol., 135: 89-99 doi:10.1007/BF01309767
. PDF(249KB)
. FPDF(win)
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Jinyan Wu
. Chenyan Hong
. Youjun Shang
. Shuanghui Yin
. Na Zhao
. Ye Jin
. Xiangtao Liu
. Qingge Xie
Related articles
. Porcine reproductive and respiratory syndrome virus (PRRSV)
. Multiplex RT-PCR
. Conventional single RT-PCR
Tools
. Email to a friend
. Post a comment


